9 Basic data visualization
9.1 Learning objectives
This session’s overall learning outcome is to:
- Describe of the “grammar” for making plots and use the ggplot2 package to make it easier to create them.
Specific objectives are to:
- Describe and list some features of the “Grammar of Graphics” approach to creating plots, and connect that to how ggplot2 works.
- Identify ways to present your figures in an output document by using Quarto chunk options.
- Explain why some commonly used graphs in science are inappropriate to use for certain data, like the barplot with mean and standard error.
- Use the
geom_point()andgeom_smooth()functions to create a scatterplot of two continuous variables, orgeom_bar()for showing counts of discrete variables. - Recognize the importance of writing readable code that follows a consistent style guide and use the styler package to help with that.
9.2 📖 Reading task: Basic principles for creating graphs
Because this is fairly short, you don’t need to spend too much time going over it.
Time: ~4 minutes
Making graphs in R is relatively easy compared to other programs and can be done with very little code. Because it takes a few lines of code to create multiple types of plots, it gives you some time to consider: why you are making them; whether the graph you’ve selected is the most appropriate for your data or results; and how you can design your graphs to be as accessible and understandable as possible.
To start, here are some tips for making a graph:
- Whenever possible or reasonable, show raw data values rather than summaries (e.g. means).
- Though commonly used in scientific papers, avoid barplots with means and error bars as they greatly misrepresent the data (we’ll cover why later).
- Use colour to highlight and enhance your message, and make the plot visually appealing.
- Use a colour-blind friendly palette to make the plot more accessible to others (more on this later too).
There are also excellent online books on this that are included in the resources page of the Guides website.
9.3 📖 Reading task: Basic structure of using ggplot2
Time: ~6 minutes
ggplot2 is an implementation of the “Grammar of Graphics” (“gg”). This is a powerful approach to creating plots because it provides a set of structured rules (a “grammar”) that allow you to expressively describe components (or “layers”) of a graph. Since you are able to describe the components, it is easier to then implement those “descriptions” in creating a graph. There are at least four aspects to using ggplot2 that relate to its “grammar”:
- Aesthetics,
aes(): How data are mapped to the plot, including what data are put on the x and y axes, and/or whether to use a colour for a variable. - Geometries,
geom_functions: Visual representation of the data, as a layer. This tells ggplot2 how the aesthetics should be visualized, including whether they should be shown as points, lines, boxes, bins, or bars. - Scales,
scale_functions: Controls the visual properties of thegeom_layers. Can be used to modify the appearance of the axes, to change the colour of dots from, e.g., red to blue, or to use a different colour palette entirely. - Themes,
theme_functions ortheme(): Directly controls all other aspects of the plot, such as the size, font, and angle of axis text, and the thickness or colour of the axis lines.
There is a massive amount of features in ggplot2. Thankfully, ggplot2 was specifically designed to make it easy to find and use its functions and settings using tab auto-completion. To demonstrate this feature, try typing out geom_ and you’ll start seeing a menu pop-up with a list of functions that start with geom_. You can then use the arrow keys to move up and down the list and then hit either Enter or Tab to select the function. You can use this with scale_ or the options inside theme(), for instance try typing out theme(axis. to see all options for the axis a list of theme settings related to the axis will pop up.
So, why do we teach ggplot2 and not base R plotting? Base R plotting functionality is quite good and you can make really nice publication-quality graphs. However, there are several major limitations to base R plots from a beginner and a user-interface perspective:
- Function and argument names are inconsistent and opaque (e.g. the
cexargument can be used to magnify text and symbols, but you can’t immediately tell from the name that it does that). - User-friendly documentation that is accessible to a broad range of people is not much of a priority, so often the help documentation isn’t written with beginners in mind.
- Graphs are built similar to painting on a canvas; make a mistake and you need to start all over (e.g. restart R).
These limitations are due to the fact that base R plotting was developed:
- By different people over different periods of time.
- By people who were/are mostly from statistics and maths backgrounds.
- By people who (generally) don’t have training in principles of software user-design, user-interface, or engineering.
- Without a strong “design philosophy” to guide development.
- During a time when auto-completion didn’t really exist or was sub-optimal, so short function and object names were more important than they are today.
On the other hand, ggplot2:
- Has excellent documentation for help and learning.
- Has a strong design philosophy that makes it easier to use.
- Works in “layers”, so you don’t have to start over if you make a mistake.
- Works very well with auto-completion.
- Uses function and argument naming that is consistent and descriptive (in plain English).
These are the reasons we teach and use ggplot2.
9.4 Plot one continuous variable
Very often you want to get a sense of your data, one variable (i.e. column in a data frame) at a time. You create plots to see the distribution of a variable and visually inspect the data for any problems. There are several ways of plotting continuous variables like age or BMI in ggplot2. For discrete variables like diabetes status, there is really only one way.
We use the word variable to mean a column in a data frame that is tidy. If you recall the definition of tidy data, it consists of “variables” (columns) and “observations” (rows) of a data frame. To us, a “variable” is something that we are interested in analyzing or visualizing, and which only contains values relevant to that measurement For instance, an age variable must only contain values in a unit of time like years.
Our post_meal_data data frame is fairly tidy, though there are a few things we will fix in later sessions. Rows are individual participants at a specific time and the columns are the variables that were measured, like weight. Let’s visually explore our data! In the LearningR project in the docs/learning.qmd file, create a new second-level header on the bottom of the file called ## Plot one continuous variable and create a new code chunk below the header with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”). Then we can make our first plot!
Since BMI is a strong risk factor for diabetes, let’s check out the distribution of BMI among the participants. There are two good geoms for examining distributions for continuous variables: geom_density() and geom_histogram(). Before we can make a plot, we need to tell ggplot2 what data we are using and which variable to put in which axis or dimension. For that we use the ggplot() function with the aes() function used inside of it.
Run this code by using Ctrl-EnterCtrl-Enter. You’ll get a blank plot. That’s because we haven’t told ggplot2 what kind of plot we want to make, which needs a geom_ function.
We’ll start with creating a histogram, which is a useful way of visualizing the distribution of a continuous variable. We do that with geom_histogram(), but you can easily replace the code with geom_density() to make a density plot. Note that it is good practice to always create a new line after the +.
docs/learning.qmd
ggplot(post_meal_data, aes(x = BMI)) +
geom_histogram()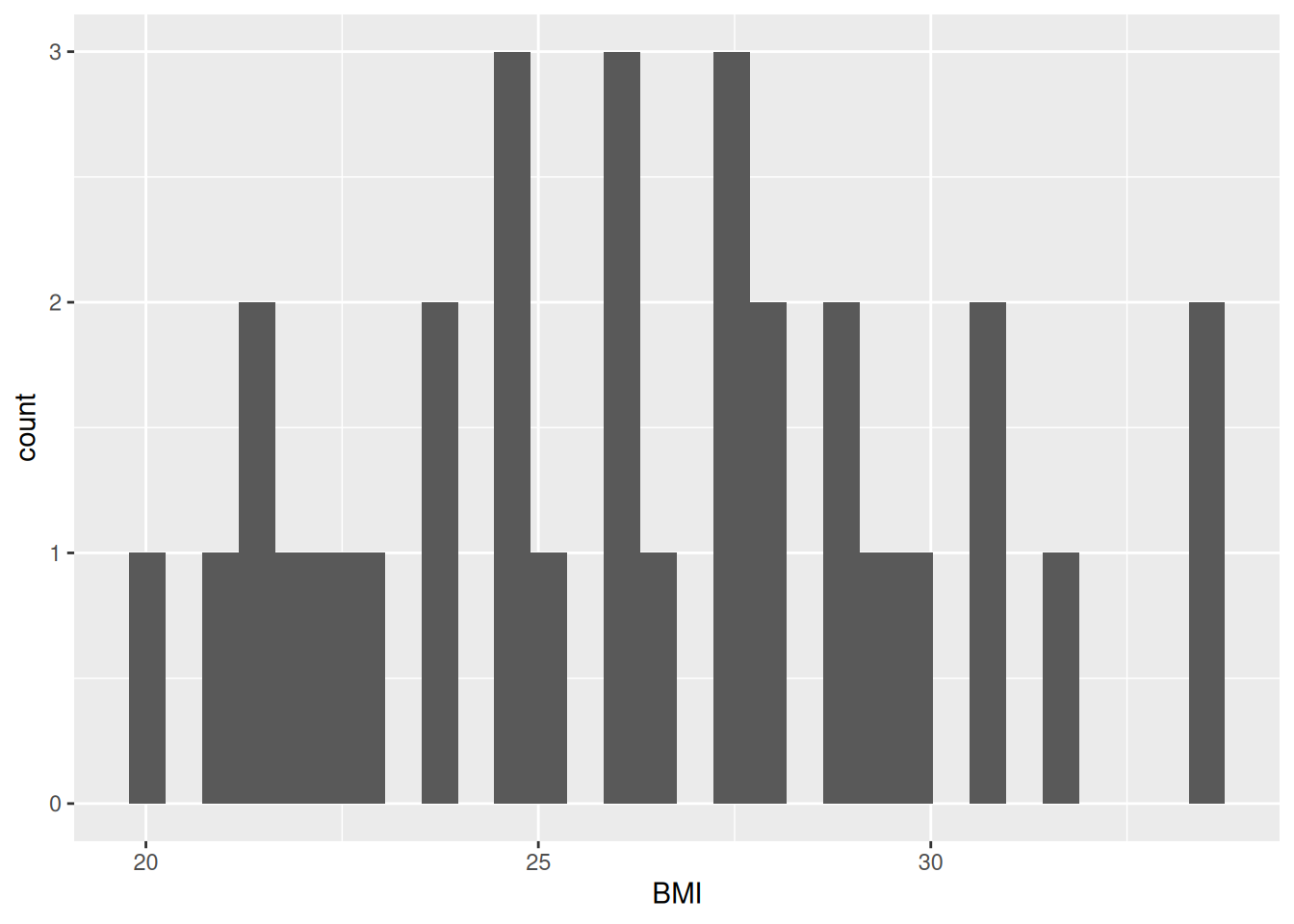
If your data has missing values, ggplot2 gives us a warning about dropping the missing values. Like many functions in R, especially the summary statistic functions like mean(), you can set the argument na.rm = TRUE in the geom_histogram() function, as well as in other geom_* functions.
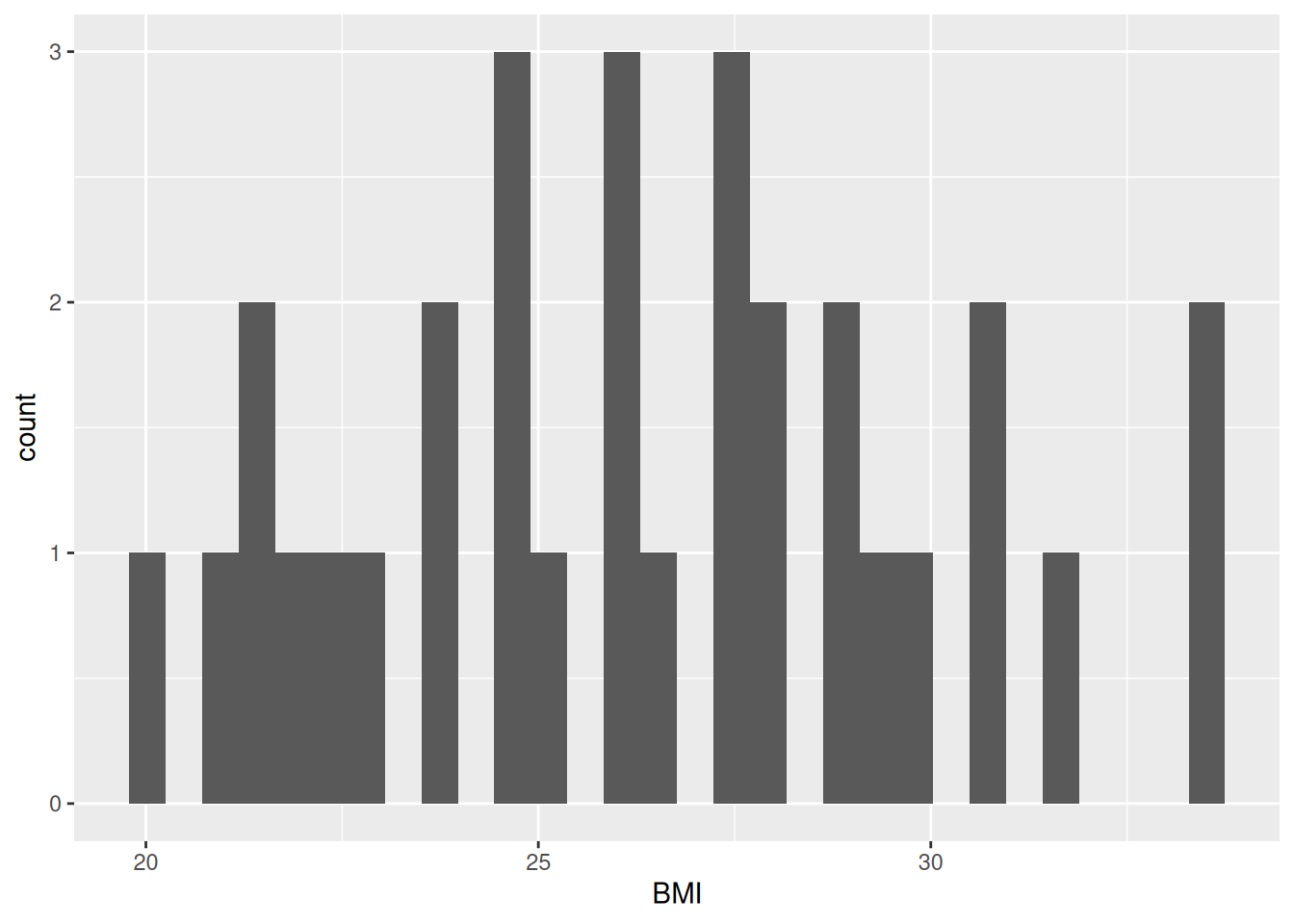
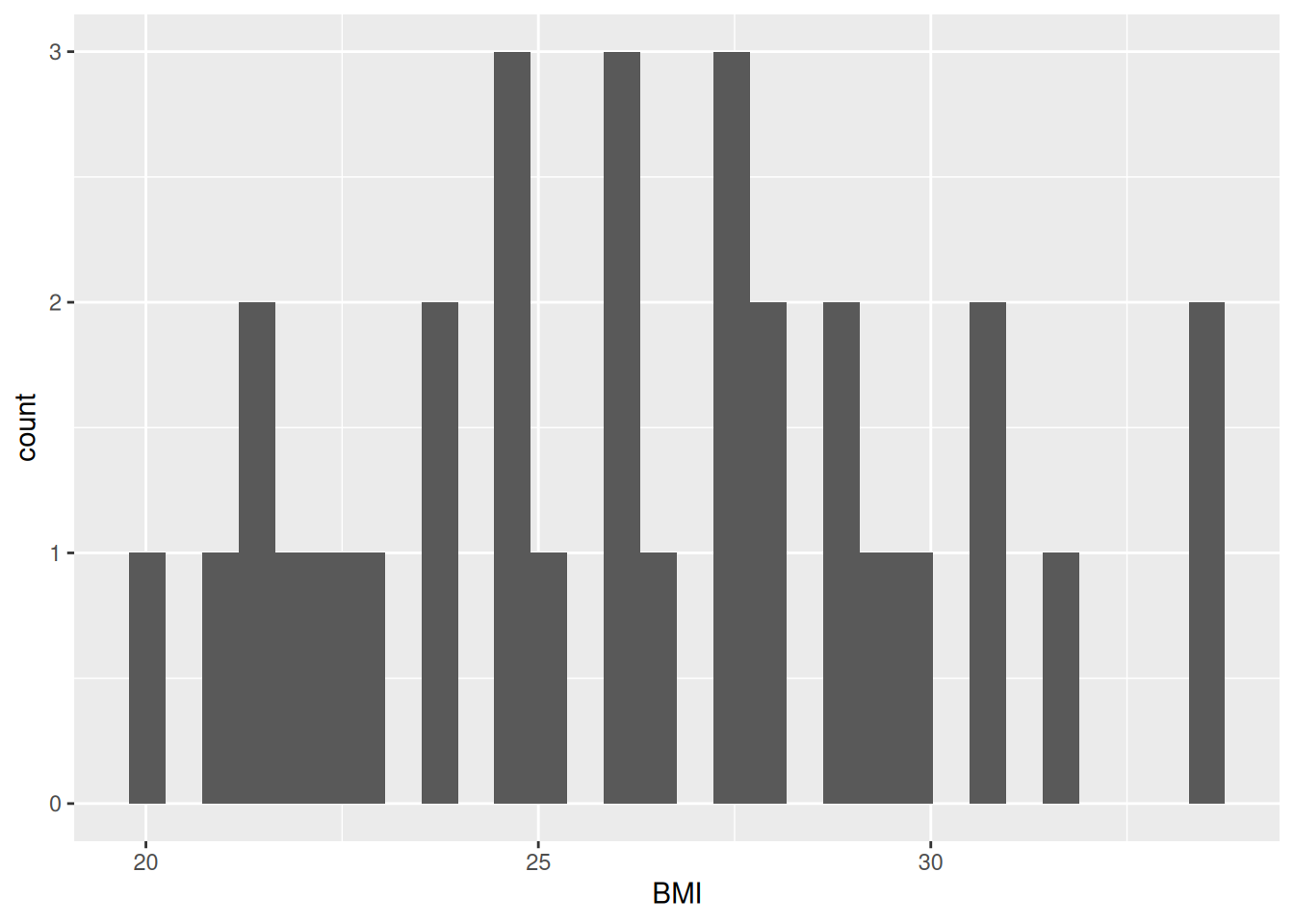
Our plot shows that, for the most part, there is a even distribution with BMI. In general, it is good practice to create a new code chunk for each plot in Quarto for several reasons. One, it makes it easier to maintain a nice readable code and, two, there are some chunk options that only work with one figure. However, it is possible to show multiple graphs for instance side-by-side, as we will do later.
In the same code chunk, we can add a caption for the plot with the option#| fig-cap. Let’s add one as well as a figure label with #| label so we can reference it in the text by using @fig-LABEL. Figure labels must always start with fig-.
docs/learning.qmd
```{r}
#| fig-cap: "Distribution of BMI."
#| label: fig-bmi-histo
ggplot(post_meal_data, aes(x = BMI)) +
geom_histogram()
```You’ll notice we use "" quotes around the caption. A good general guideline is to use quotes for things that are natural languages, like English.
Now when we reference the figure in the text, we can use @fig-bmi-histo, to look like this: Figure 9.1.
Before we move on, let’s add and commit the changes we’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”).
9.5 🧑💻 Exercise: Plot a discrete variable
Time: ~10 minutes.
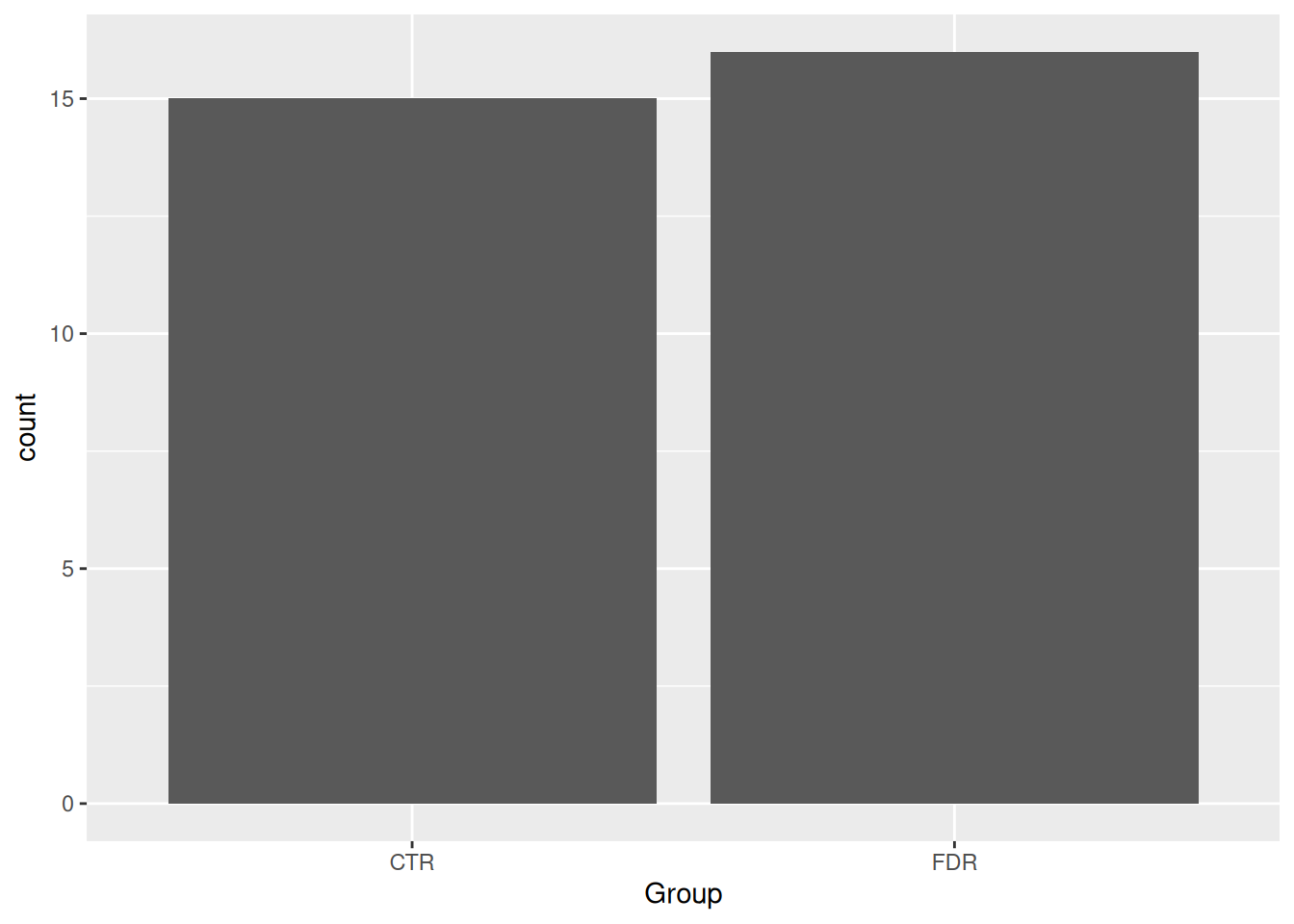
Using the same structure as the code from above, let’s plot a discrete variable. The geom_histogram() from above is appropriate for plotting continuous variables, but not for discrete variables. Sadly, there’s really only one: geom_bar(). This isn’t a geom for a barplot though; instead, it shows the counts of a discrete variable. There aren’t too many discrete variables our dataset, only Group and glykemi, so let’s use this geom to visualize those.
For this exercise, create a new header at the bottom of the docs/learning.qmd file called ## Exercise: discrete plots. Then, copy the below code chunk template and paste it below the header. Complete the tasks below on this template.
docs/learning.qmd
```{r}
#| fig-cap: "___"
#| label: fig-___
ggplot(post_meal_data, aes(x = ___)) +
___()
```
See @fig-___ above for a cool plot!- Decide on either
Grouporglykemito plot on thexaxis in theaes()function, replacing the___with the one you chose. - Replace the
___below theggplot()function withgeom_bar(). - Add a caption to the plot with
#| fig-capwhere the___is placed. - Add a label in the
#| labeloption section, again, replacing thefig-___with a label that describes the plot, likefig-group-bar - Replace the
___under the code chunk where the@fig-reference is with the label you chose in the plot (like@fig-group-bar). - Render the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”) to see the plot you created.
- Add and commit the changes you’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”).
Click for the solution. Only click if you are struggling or are out of time.
```{r solution-discrete-variables}
#| eval: false
#| code-fold: true
#| code-summary: '**Click for the solution**. Only click if you are struggling or are out of time.'
# This is a potential solution
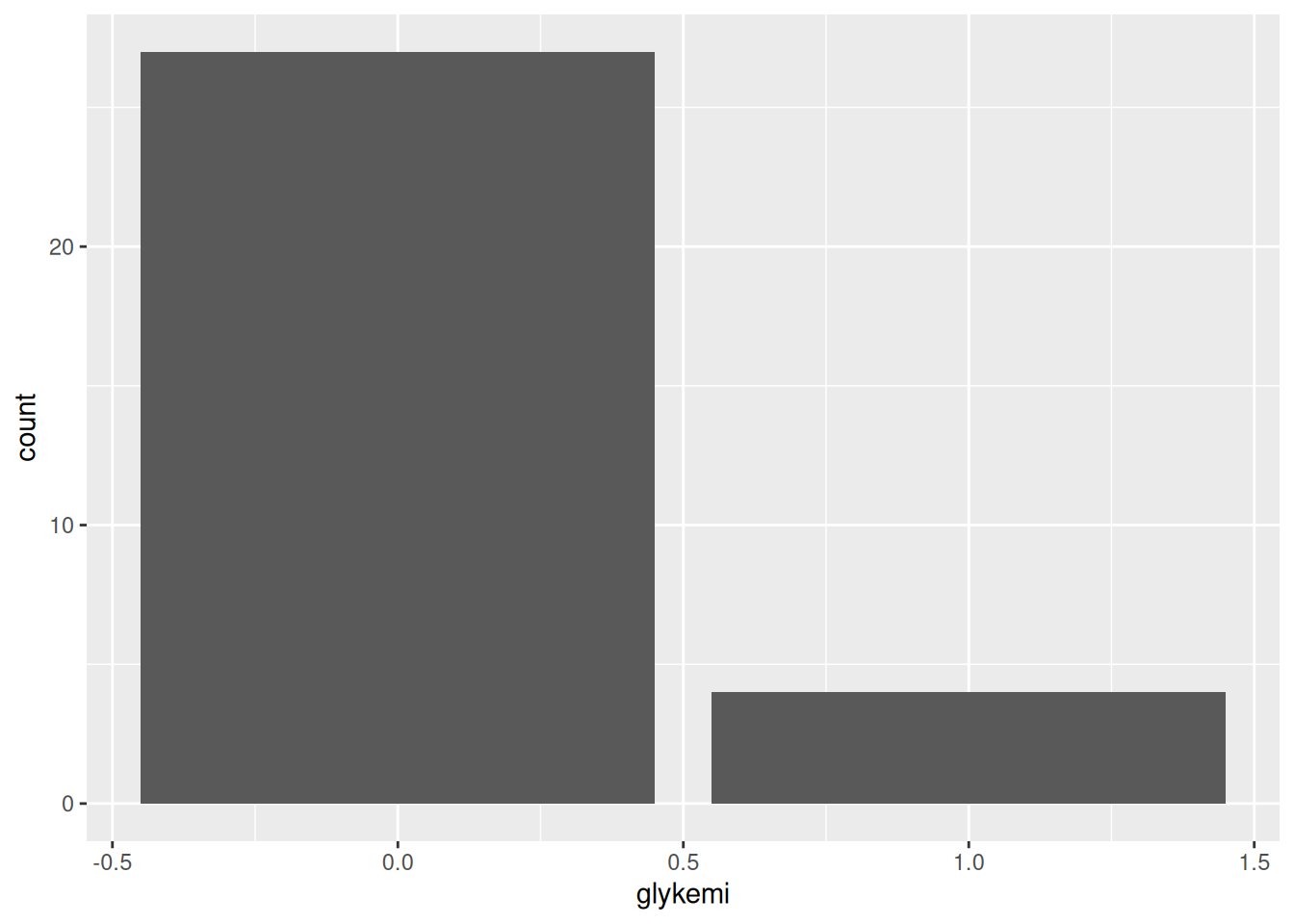
#| fig-cap: "Distribution of glykemi."
#| label: fig-glykemi-bar
ggplot(post_meal_data, aes(x = glykemi)) +
geom_bar()
```9.6 Plot two discrete variables
Unlike plotting continuous variables, there are not as many plotting options for discrete variables, even two of them, without major data wrangling. You’ve already tried the geom_bar() for one variable, let’s do one for two variables. We can use the geom_bar() “fill” option to have a certain colour for different levels of a variable. Let’s use this to see differences between Group and glykemi. Create a new header at the bottom of the file called ## Plotting two discrete variables and create a new code chunk below that with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”). While we could put either Group or glykemi on the x-axis, let’s put Group on the x-axis and glykemi as the fill.
Run this code with Ctrl-EnterCtrl-Enter. You’ll see we get a warning message. That’s because the glykemi variable is actually a number, and ggplot2 needs a character vector for fill. We can easily fix this by using as.character() on the glykemi variable in the aes() function.
docs/learning.qmd
ggplot(post_meal_data, aes(x = Group, fill = as.character(glykemi))) +
geom_bar()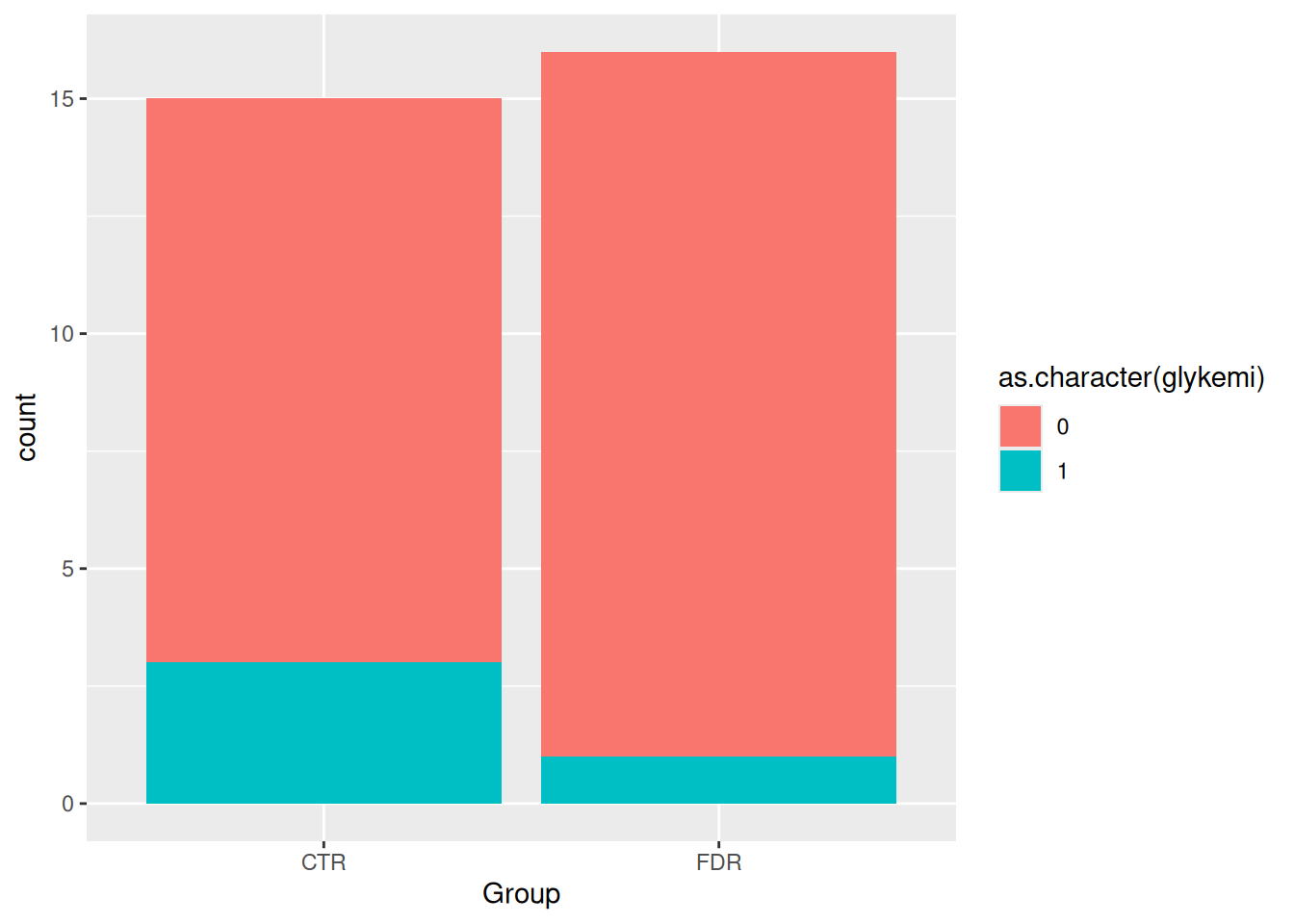
Running this code with Ctrl-EnterCtrl-Enter will show you a barplot of Group with glykemi as the fill, without the warning message. But the legend on the side is a bit ugly. It’s easy to fix with the use of the theme() function, but we won’t cover that in this session.
By default, geom_bar() will make “fill” groups stacked on top of each other. In this case, it isn’t really that useful, so let’s change them to be sitting side by side. For that, we need to use the position argument with a function called position_dodge(). This new function takes the “fill” grouping variable and “dodges” them (moves them) to be side by side.
docs/learning.qmd
ggplot(post_meal_data, aes(x = Group, fill = as.character(glykemi))) +
geom_bar(position = position_dodge())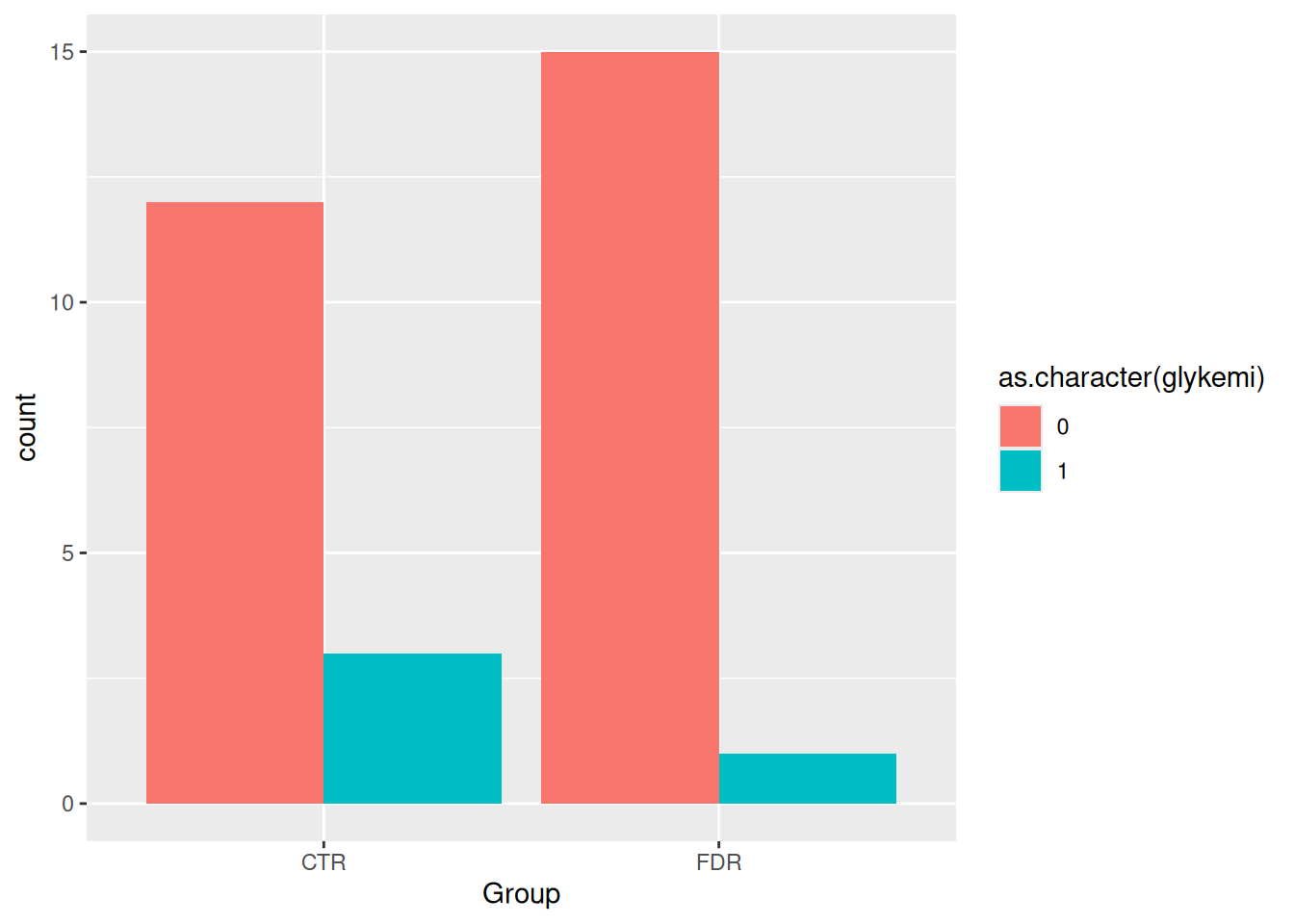
We can see from this plot there are very few people with the glykemi as 1. Let’s render the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”) to see the plot in the output document, then add and commit the changes you’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”).
9.7 Putting two plots side by side
While we said in general you should use one figure per code chunk, if you want figures to be side by side as “one figure”, you can use a feature in Quarto to place figures into other different layouts. It is described in more detail in Quarto’s Figures page.
First, let’s create a new header called ## Side by side plots and create a new code chunk with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”) below that. Then write out the same code for the figures from above.
Before we continue, let’s see what that looks like by rendering the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”). See how the figures are placed one after the other?
Using Quarto, let’s layout these two figures so they are side by side, with sub-captions below each figure, for when we render the output document.
We can layout the figures using the #| layout-ncol (or #| layout-nrow or #| layout) option. We can then combine it with captions and sub-captions using #| fig-subcap to have a nice output!
```{r side-by-side-figs}
#| label: fig-bmi-glycemia
#| fig-cap: "BMI and glycemia, side by side."
#| fig-subcap:
#| - "Distribution of BMI."
#| - "Number of those with glycemia."
#| layout-ncol: 2
ggplot(post_meal_data, aes(x = BMI)) +
geom_histogram()
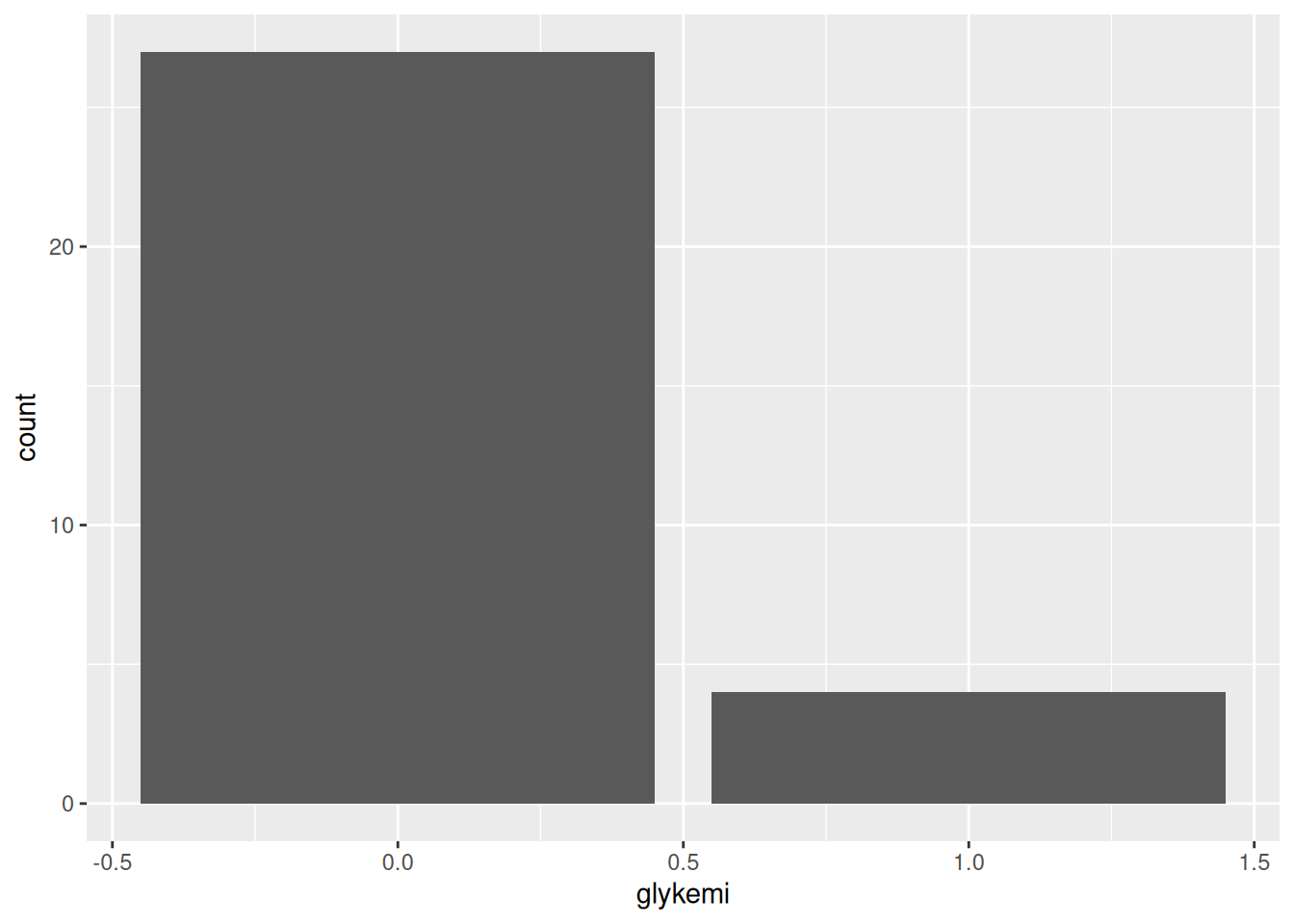
ggplot(post_meal_data, aes(x = glykemi)) +
geom_bar()
```Render the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”) to see what Figure 9.2 looks like! Neat eh 😀
Before we move on, let’s add and commit the changes we’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”).
9.8 📖 Reading task: Be careful with certain plots
Time: ~5 minutes
Before continuing with plotting, let’s take a minute to talk about a commonly used barplots with mean and error bars. In all cases, barplots should only be used for discrete (categorical) data where you want to show counts or proportions. As a general rule, they should not be used for continuous data. This is because the commonly used “bar plot of means with error bars” actually hides the underlying distribution of the data. To have a better explanation of this, you can read the article on why to avoid barplots after the course. The image below was taken from that paper, and briefly demonstrates why this plot type is not useful.
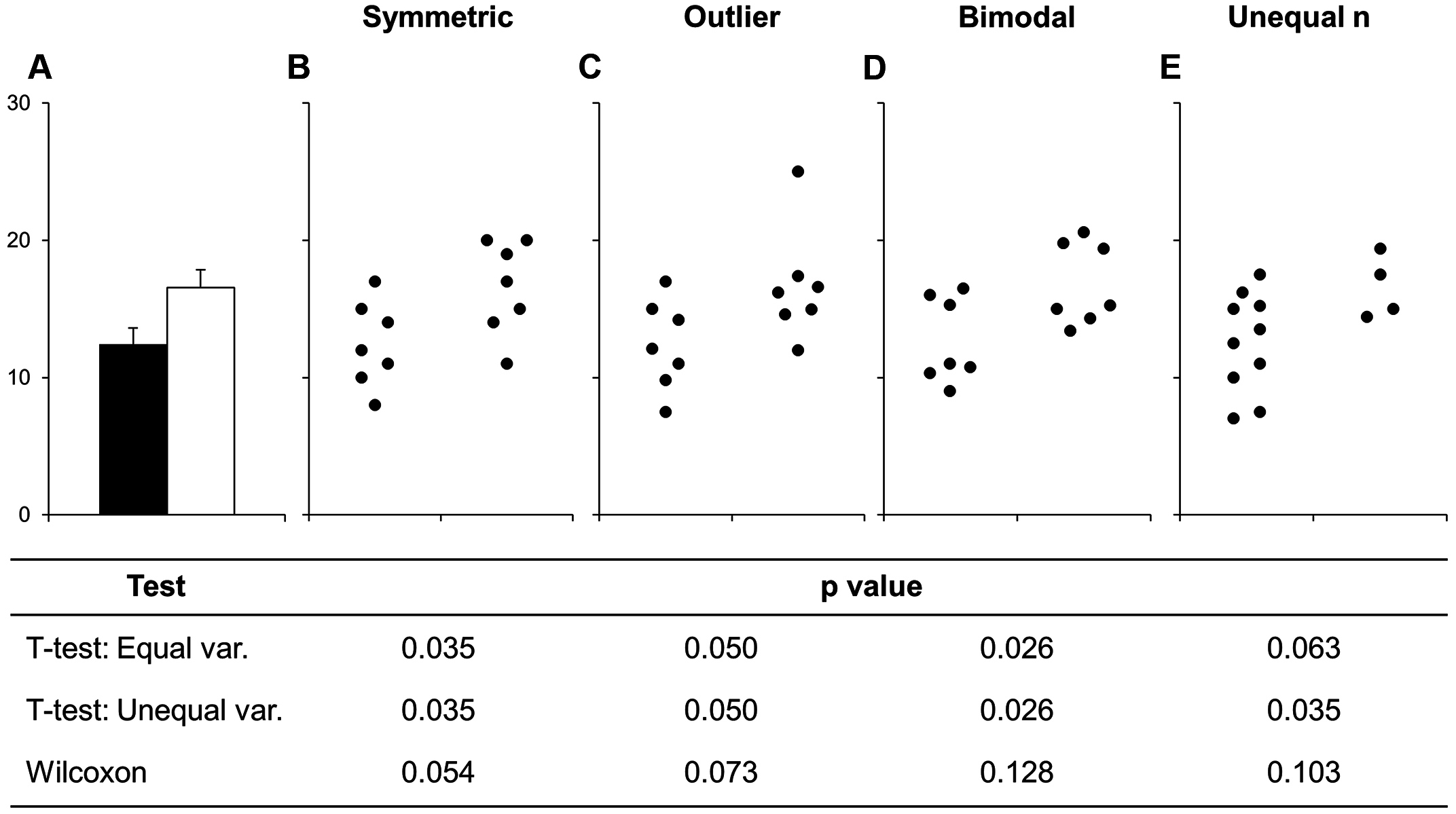
If you do want to create a barplot, you’ll quickly find out that it is actually quite hard to do in ggplot2. The reason it is difficult to create in ggplot2 is by design: it’s a bad plot to use, so use something else.
9.9 📖 Reading task: Making code more readable
Time: ~6 minutes
Now that we’ve started writing some code, we should talk about how we should write code. We write code not for the computer, but for ourselves in the future and for others. This means that code should be written in a way that is readable and relatively understandable, at least to someone with some knowledge of R.
Let’s take a look at some code that is in some way either wrong or very poorly written. What are some things that could be improved or are just wrong? You don’t necessarily need to understand what the code does, but rather, how readable it is.
# Object names
DayOne
T <- FALSE
c <- 9
# Spacing
x[,1]
x[ ,1]
mean (x, na.rm = TRUE)
mean( x, na.rm = TRUE )
height<-feet*12+inches
df $ z
x <- 1 : 10
# Tabs
if (x == 1) {
y <- 2
}These issues can actually be broken down into two categories: naming and styling.
Naming issues
This issue is difficult to fix automatically by the computer, because it usually needs a human with the context knowledge in order to fix. Fixing it properly comes with experience and knowledge. Based on naming issues, the code above has two types of issues:
Syntax issues: The
T <- FALSEis wrong becauseTalready exists and is a short hand forTRUEwhilec <- 9is wrong becausecis already the name of the functionc(). You normally don’t want to name code based on something that already exists in base R (“naming conflicts” between packages is fine though, since there are ways to identify and fix that). These issues can only be fixed manually.Semantic issues: The
DayOneobject is not descriptive. Day one of what? Also,DayOneis in the format ofPascalCaseand in general,snake_case(all lowercase with underscores) has been found to be better for readability. And even ifTandcweren’t syntax issues, they are not descriptive. As you learn more R, you will find a lot of functions with very short names, or names that are not descriptive at all. In the past, there were no tab-completion tools, so typing out long names was painful. Now we have powerful auto-completion tools, which means you don’t have the restriction for shorter names. You should use always write descriptive, plain language names instead of short, concise ones. Stick to a style guide.
Styling issues
Even though R doesn’t care about naming, spacing, and indenting, it really matters how your code looks. Coding is just like writing. Even though you may go through a brainstorming note-taking stage of writing, you eventually need to write correctly so others can read and understand what you are trying to say. In coding, brainstorming is fine, but eventually you need to code in a readable way. That’s why using a style guide is really important.
This issue is much easier to fix automatically. Most of these issues come down to effective or correct use of spacing and tabs.
- Spacing: The spacing around the brackets, commas, and math are inconsistent. Spacing is one of the most common stylistic issues in code, as the code will still run, but it is harder to read.
- Tabs: Tabs are used to indent code to make it easier to scan and read. You tab code within functions, if statements, and other situations to make it easier to read.
9.10 Using styler to fix code styling issues
To fix the styling issues, we can use a package called styler, which follows the tidyverse style guide, to automatically reformat our code into the correct style. With styler you can style multiple files at once, one file at a time, or based on code you select and highlight. We will style our files a lot as we write code throughout this course. We’ll use it with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “style file”), which should show the “Style active file” option.
If you are using RStudio version at least 2024.12.0, you can set up styler to run automatically when you save a file. Go to “Tools”, “Global Options”, click the “Code” section, then click the “Formatting” pane, and finally select “styler” from the drop-down menu list. Click ok and now, whenever save your file, it will re-format it for you.
The thing to note, is that styler isn’t perfect and can’t fix naming issues or issues that require human consideration. For example, it can’t change objects that are named T or c to something else. But styler is a good starting point to manually fixing up your code.
Let’s run styler on the docs/learning.qmd file with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “style file”). It make not make too many fixes since we don’t have too much code yet, but we’ll slow build up the habit to style our code regularly. Add and commit the changes you’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”) before we move on.
9.11 Plotting two continuous variables
There are many more types of “geoms” to use when plotting two variables. Your choice of which one to use depends on what you are trying to show or communicate, and the nature of the data. Usually, the variable that you “control or influence” (the independent variable) in an experimental setting goes on the x-axis, and the variable that “responds” (the dependent variable) goes on the y-axis.
For now, let’s focus on plotting continuous data, since our data has a lot of continuous data and often in research we work with more continuous than discrete variables. When you have two continuous variables, some geoms to use are:
-
geom_point(), which is used to create a standard scatterplot. Since it is so commonly used, we’ll use this one. -
geom_hex(), which is used to replacegeom_point()when your data are massive and creating points for each value takes too long to plot. -
geom_smooth(), which applies a “regression-type” line to the data.
Let’s check out how BMI may influence the area under the curve for blood glucose after the meal using a point plot. The area under the curve of glucose is a measure of how much glucose is present in the blood over a fixed period of time. A higher area under the curve for glucose usually suggests that the body has a harder time handling glucose. But, like everything in biology, it’s more complex than that. But we won’t cover that complexity in this course.
First, write out a new Markdown header called ## Plotting two continuous variables at the bottom of the document and create a new code chunk below the header with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”). Like with the previous plot we created using BMI, we’ll put BMI on the x axis in the aes() function, since we are assuming that BMI “causes” or contributes to higher glucose in the blood after a meal. Then we put the area under the curve for glucose (auc_pg) on the y axis, since it will be “responding to” or “caused by” BMI.
docs/learning.qmd
ggplot(post_meal_data, aes(x = BMI, y = auc_pg)) +
geom_point()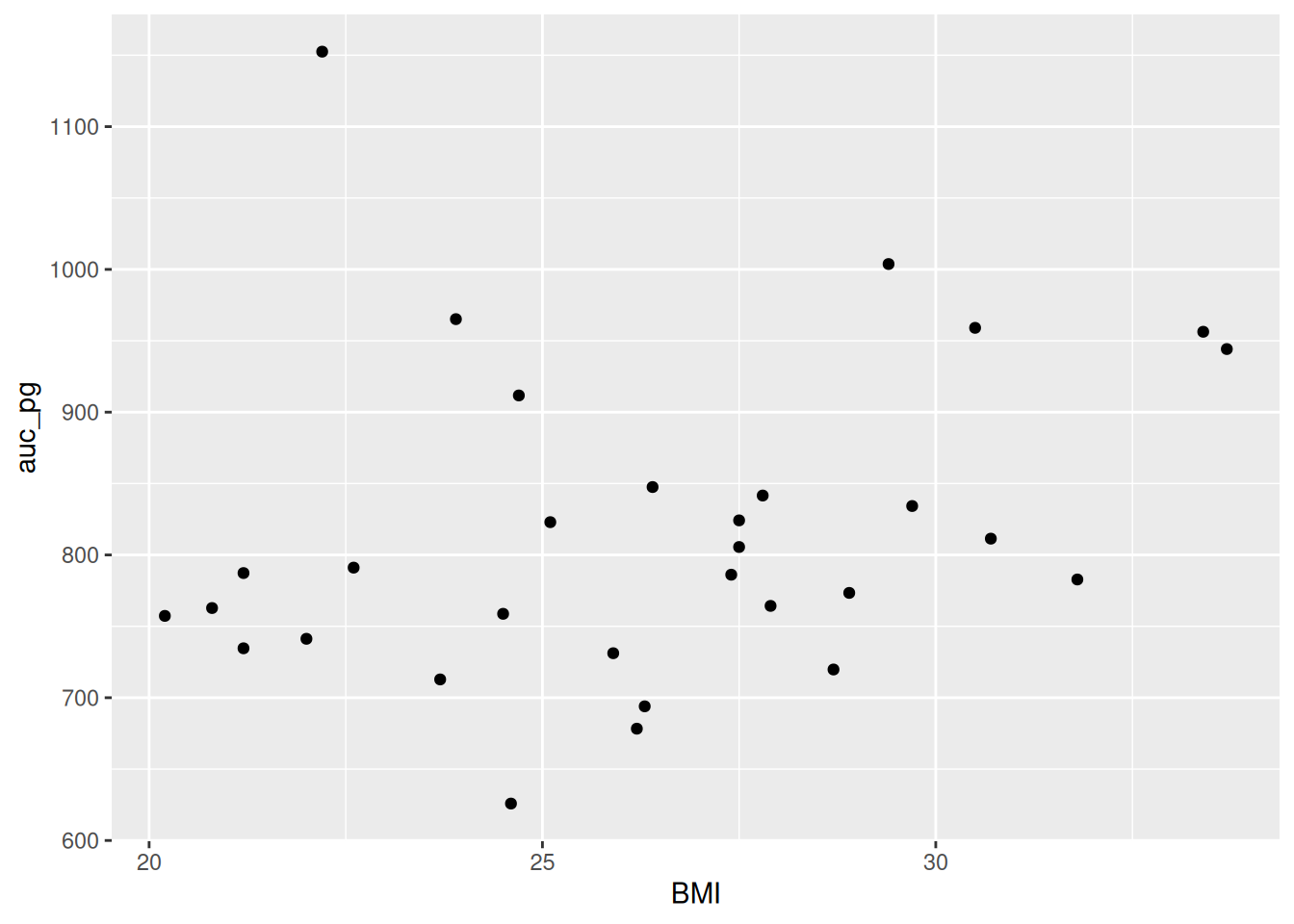
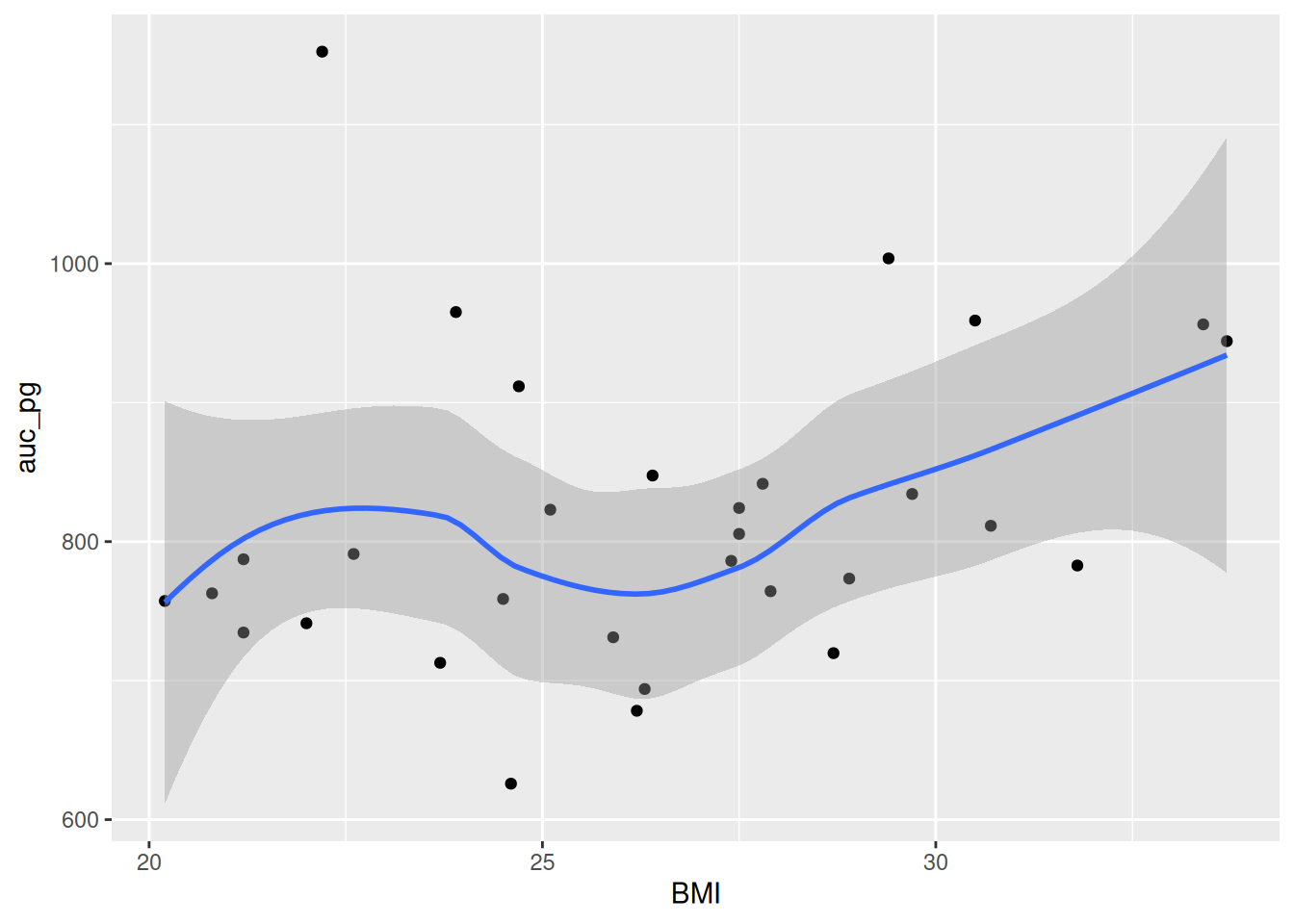
Run this code chunk with Ctrl-EnterCtrl-Enter. You’ll see a scatterplot of BMI and the area under the curve for glucose. Notice how there is a bit of an increase in glucose as BMI increases? Because ggplot2 works in layers, we can add another layer to the plot by adding a + after the geom_point() function. Let’s add a smoothing line to the plot to see if there is a general trend between BMI and glucose by using geom_smooth().
docs/learning.qmd
ggplot(post_meal_data, aes(x = BMI, y = auc_pg)) +
geom_point() +
geom_smooth()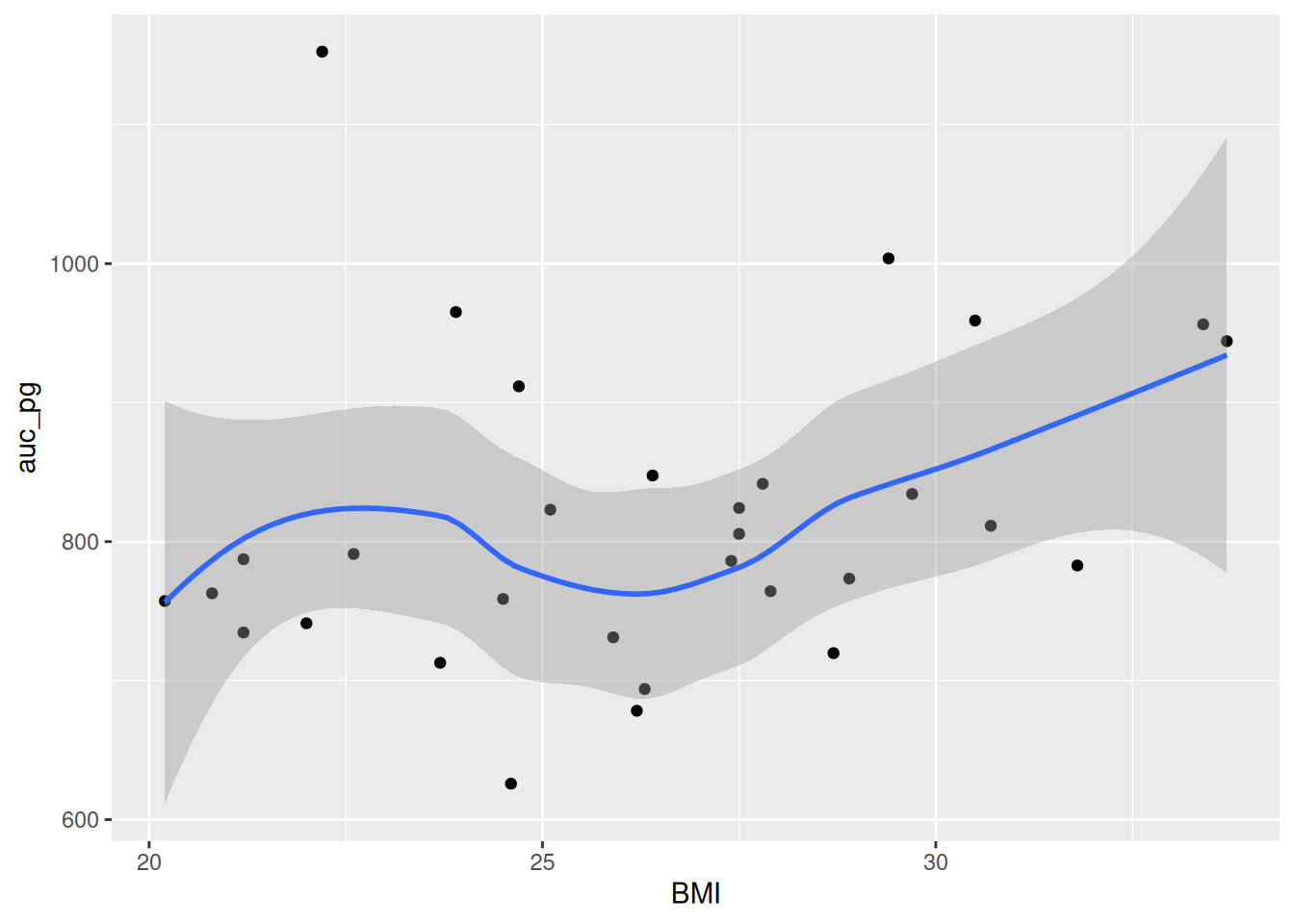
Run this code chunk with Ctrl-EnterCtrl-Enter and we’ll see a scatterplot of BMI and the area under the curve for glucose with a smoothing line on top of the points. To help build the habit of styling the code, run styler on the file with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “style file”). Before rendering our HTML document, let’s add a caption and label to the plot:
```{r}
#| fig-cap: "Scatterplot of BMI and the area under the curve for glucose."
#| label: fig-bmi-auc-pg
ggplot(post_meal_data, aes(x = BMI, y = auc_pg)) +
geom_point() +
geom_smooth()
```Now, let’s make our HTML document by rendering the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”). Now you can see the plot in the rendered document! This makes a nice smoothing line through the data and gives us an idea of general trends or relationships between the two variables.
Lastly, let’s add and commit the changes you’ve made into the Git history with Ctrl-Alt-MCtrl-Alt-M or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “commit”).
9.12 Summary
- Use a consistent style guide for code for using styler.
- Use the “Grammar of Graphics” approach together with the ggplot2 package within tidyverse to plot your data.
- Prioritize plotting raw data instead of summaries whenever possible.
-
ggplot2 has 4 levels of grammar:
aes()to say which data to plot and how,geom_to set the kind of plot to use,scale_to make the plot prettier, andtheme()to control the specifics of the plot. We covered theaes()andgeom_layers. - Only use barplots for discrete values. If applying them on continuous variables, it hides the distribution of the data.
- Use
geom_point()to create a scatterplot of two continuous variables. - Use
geom_smooth()to add a smoothing line to the scatterplot. - Use
geom_bar()to create a barplot of discrete variables.